🧮 Building a mini optimizer library#
In the following example an optax-like mini-library is built using pytreeclass. The optimizer library is then used to train a simple neural network.
[1]:
!pip install pytreeclass --quiet
Imports#
[2]:
import jax
import jax.numpy as jnp
import pytreeclass as tc
from typing import Any, TypeVar, Generic, Callable
import abc
import matplotlib.pyplot as plt
PyTree = Any
T = TypeVar("T")
Template#
[3]:
class GradientTransformation(tc.TreeClass, Generic[T]):
# TreeClass inherits from `abc.ABC`
# following the same pattern as `optax` optimizers
# https://github.com/deepmind/optax/blob/fc5de3d3951c4dfd87513c6426e30baf505d89ae/optax/_src/base.py#L85
@abc.abstractclassmethod
def update(self, _: T) -> tuple[T, "GradientTransformation"]:
pass
Optimizer functions#
[4]:
def moment_update(grads, moments, *, beta: float, order: int):
def moment_step(grad, moment):
return beta * moment + (1 - beta) * (grad**order)
return jax.tree_map(moment_step, grads, moments)
def debias_update(moments, *, beta: float, count: int):
def debias_step(moment):
return moment / (1 - beta**count)
return jax.tree_map(debias_step, moments)
def ema(decay_rate: float, debias: float = True) -> GradientTransformation:
"""Exponential moving average
Args:
decay_rate: The decay rate of the moving average.
debias: Whether to debias the moving average.
"""
class EMA(GradientTransformation):
def __init__(self, tree):
self.state = jax.tree_map(jnp.zeros_like, tree)
self.count = 0
def _update(self, grads: T) -> T:
self.count += 1
self.state = moment_update(grads, self.state, beta=decay_rate, order=1)
if debias:
return debias_update(self.state, beta=decay_rate, count=self.count)
return self.state
def update(self, grads: T) -> tuple[T, "EMA"]:
return self.at["_update"](grads)
return EMA
def adam(
*,
beta1: float = 0.9,
beta2: float = 0.999,
eps: float = 1e-8,
) -> GradientTransformation:
"""Adam optimizer
Args:
beta1: The decay rate of the first moment.
beta2: The decay rate of the second moment.
eps: A small value to prevent division by zero.
Note:
Kingma et al, 2014: https://arxiv.org/abs/1412.6980
"""
class Adam(GradientTransformation):
def __init__(self, tree):
self.mu = jax.tree_map(jnp.zeros_like, tree)
self.nu = jax.tree_map(jnp.zeros_like, tree)
self.count = 0
def _update(self, grads: T) -> T:
self.count += 1
self.mu = moment_update(grads, self.mu, beta=beta1, order=1)
self.nu = moment_update(grads, self.nu, beta=beta2, order=2)
mu_hat = debias_update(self.mu, beta=beta1, count=self.count)
nu_hat = debias_update(self.nu, beta=beta2, count=self.count)
return jax.tree_map(
lambda mu, nu: mu / (jnp.sqrt(nu) + eps), mu_hat, nu_hat
)
def update(self, grads: T) -> tuple[T, "Adam"]:
# since self._update mutates the state, we need to use self.at
# to return the method value and the mutated state
return self.at["_update"](grads)
return Adam
def scale(rate_func: Callable[[int], float]) -> GradientTransformation:
"""Scale the gradients by a scheduler function"""
class Scale(GradientTransformation):
def __init__(self, _=None):
self.count = 0
self.rate = rate_func(self.count)
def _update(self, grads: T) -> T:
self.count += 1
self.rate = rate_func(self.count)
return jax.tree_map(lambda x: x * self.rate, grads)
def update(self, grads: T) -> tuple[T, "Scale"]:
return self.at["_update"](grads)
return Scale
def chain(
*transformations: tuple[GradientTransformation, ...]
) -> GradientTransformation:
"""Chain multiple transformations together similar to `optax.chain`"""
class Chain(GradientTransformation):
def __init__(self, tree):
self.transformations = tuple(T(tree) for T in transformations)
def _update(self, grads: T) -> T:
state = []
for transformation in self.transformations:
grads, optim_state = transformation.update(grads)
state += [optim_state]
self.transformations = tuple(state)
return grads
def update(self, grads: T) -> tuple[T, "Chain"]:
return self.at["_update"](grads)
return Chain
Construct a fully connected neural network#
[5]:
class FNN(tc.TreeClass):
def __init__(self):
self.w1 = jax.random.normal(jax.random.PRNGKey(0), [1, 10])
self.b1 = jnp.zeros([10], dtype=jnp.float32)
self.w2 = jax.random.normal(jax.random.PRNGKey(1), [10, 1])
self.b2 = jnp.zeros([1], dtype=jnp.float32)
def __call__(self, x: jax.Array) -> jax.Array:
x = x @ self.w1 + self.b1
x = jax.nn.relu(x)
x = x @ self.w2 + self.b2
return x
Train function#
[6]:
def scheduler(count: int) -> float:
# build a scheduler function
return jnp.where(count < 2_000, -1e-2, jnp.where(count < 5_000, -1e-3, -1e-4))
fnn = FNN()
optim = chain(adam(), scale(scheduler))
optim_state = optim(fnn)
def loss_func(fnn: FNN, x: jax.Array, y: jax.Array) -> jax.Array:
return jnp.mean((fnn(x) - y) ** 2)
@jax.jit
def train_step(fnn, optim_state, x, y):
grads = jax.grad(loss_func)(fnn, x, y)
grads, optim_state = optim_state.update(grads)
fnn = jax.tree_map(lambda p, g: p + g, fnn, grads)
return fnn, optim_state
x = jnp.linspace(-1, 1, 100).reshape(-1, 1)
y = x**2 + 0.1
for i in range(1, 10_000 + 1):
fnn, optim_state = train_step(fnn, optim_state, x, y)
if i % 1_000 == 0:
loss = loss_func(fnn, x, y)
learning_rate = optim_state.transformations[1].rate
print(
f"Epoch={i:003d}\tLoss: {loss:.3e}\t Gradient scaler: {learning_rate:.3e}"
)
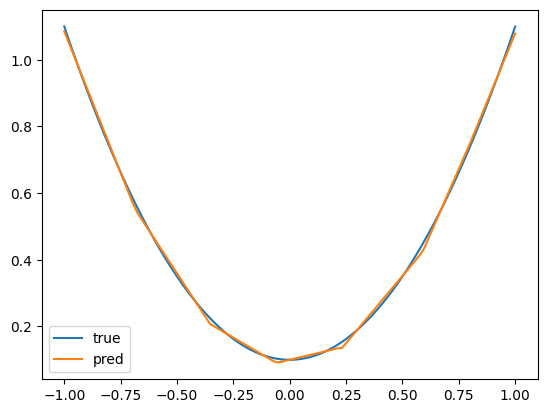
plt.plot(x, y, label="true")
plt.plot(x, fnn(x), label="pred")
plt.legend()
Epoch=1000 Loss: 1.322e-04 Gradient scaler: -1.000e-02
Epoch=2000 Loss: 8.230e-05 Gradient scaler: -1.000e-03
Epoch=3000 Loss: 8.062e-05 Gradient scaler: -1.000e-03
Epoch=4000 Loss: 7.908e-05 Gradient scaler: -1.000e-03
Epoch=5000 Loss: 7.730e-05 Gradient scaler: -1.000e-04
Epoch=6000 Loss: 7.702e-05 Gradient scaler: -1.000e-04
Epoch=7000 Loss: 7.657e-05 Gradient scaler: -1.000e-04
Epoch=8000 Loss: 7.584e-05 Gradient scaler: -1.000e-04
Epoch=9000 Loss: 7.464e-05 Gradient scaler: -1.000e-04
Epoch=10000 Loss: 7.271e-05 Gradient scaler: -1.000e-04
[6]:
<matplotlib.legend.Legend at 0x1313c18d0>